Kidney Tumor Segmentation using nnUNet
A Capstone Project Journey with RMIT University and IMRA Surgical
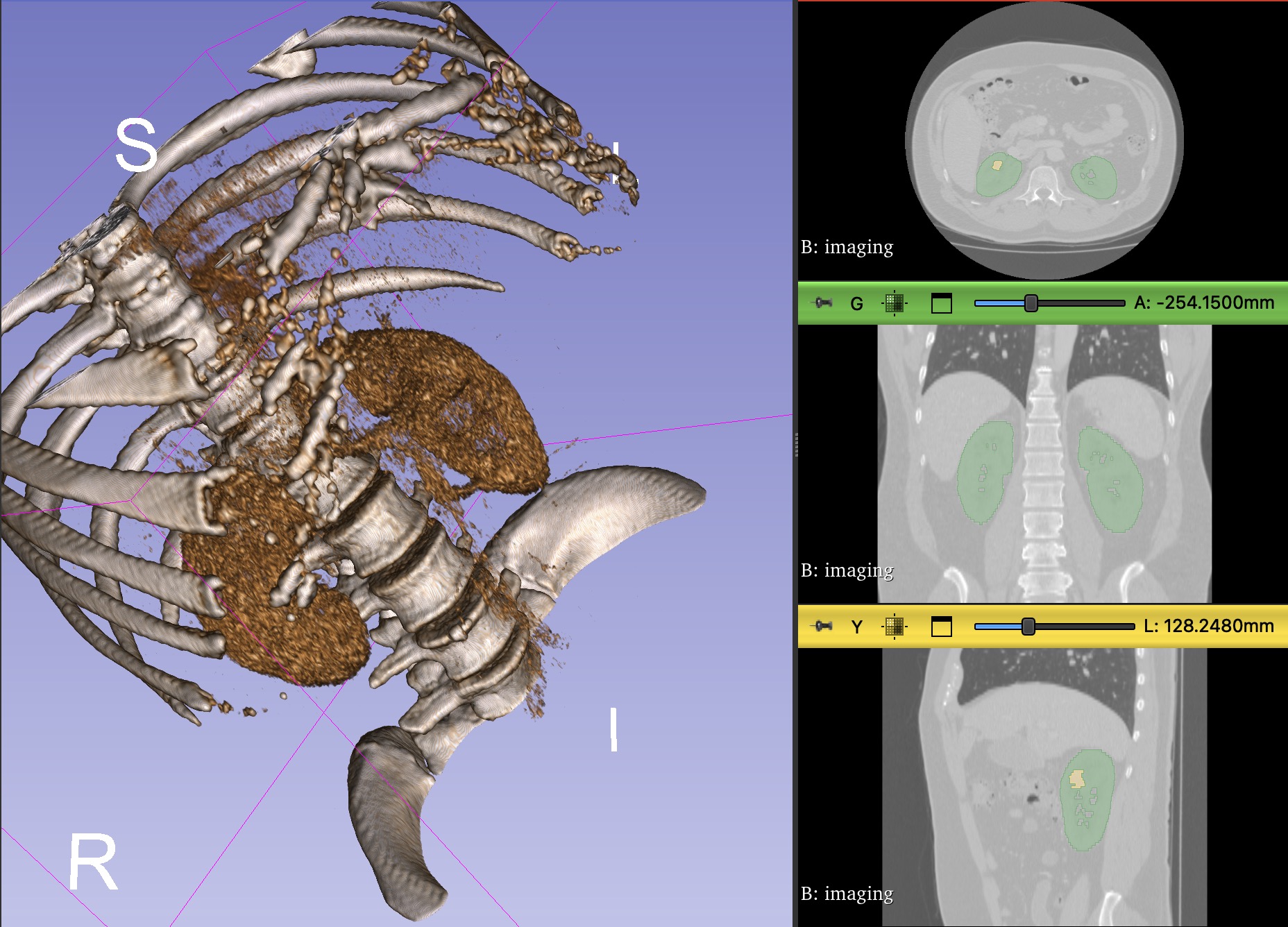
The Challenge
When I embarked on this capstone project at the International Medical Robotics Academy (IMRA), I knew I was in for an exciting and challenging experience. The goal was to develop a machine learning model that could automate the process of human organ modelling from patient CT scans, specifically focusing on kidney tumor segmentation. Little did I know, this project would not only test my technical skills but also teach me valuable lessons about collaboration, perseverance, and the power of innovation in healthcare.
The IMRA Team
One of the highlights of this project was the incredible team at IMRA. Led by the visionary founder, Professor Tony Costello, and supported by dedicated staff members like Andrew, Adam, Shaun, and Grace, their passion for medical innovation was truly inspiring. Their guidance and expertise throughout the project were invaluable, and I felt privileged to be a part of such a talented and driven team.
Project Overview
The primary objective of this project was to streamline the creation of synthetic tissue models for surgical training. By automating the segmentation of kidney tumors from CT scans, we aimed to reduce the time and effort required for manual labeling, enabling efficient generation of 3D printable models. This would ultimately enhance the training experience for surgeons and contribute to improved patient outcomes.
Understanding CT Scans and DICOM Data
Before diving into the technical aspects of the project, let's briefly understand what CT scans and DICOM data are. A CT (Computed Tomography) scan is a medical imaging technique that uses X-rays to create detailed cross-sectional images of the body. These scans provide valuable information about internal organs, bones, and tissues.
DICOM (Digital Imaging and Communications in Medicine) is a standard format used for storing and transmitting medical images, including CT scans. DICOM files contain not only the image data but also metadata such as patient information, scan parameters, and image dimensions.
Exploring 3D Slicer
To better understand the anatomy and structure of the kidneys and tumors, I utilized a powerful open-source software called 3D Slicer. This software allowed me to visualize and interact with the DICOM data in three dimensions. With 3D Slicer, I could view the CT scans from different angles, adjust contrast and brightness, and even perform manual segmentation to identify the regions of interest.
Using 3D Slicer provided valuable insights into the challenges of manual segmentation and highlighted the need for automated methods. It also helped me gain a deeper understanding of the anatomical structures and the variations in kidney and tumor appearance across different patients.
Understanding Image Segmentation
Image segmentation is a crucial technique in medical image analysis that involves partitioning an image into multiple segments or regions of interest. In the context of our project, image segmentation played a vital role in identifying and delineating the boundaries of kidneys and tumors from CT scans.
Types of Image Segmentation
There are various approaches to image segmentation, each with its own strengths and limitations. Some common techniques include:
- Thresholding: This method involves separating an image into different regions based on pixel intensity values. It is simple and computationally efficient but may not always capture complex structures accurately.
- Region Growing: Starting from a seed point, this technique iteratively expands the segmented region by including neighboring pixels that meet certain similarity criteria. It can handle irregular shapes but may be sensitive to noise and initial seed placement.
- Edge Detection: This approach identifies edges or boundaries between regions based on sharp changes in pixel intensities. It is effective in delineating object boundaries but may struggle with weak or discontinuous edges.
- Clustering: Clustering algorithms, such as K-means or Gaussian Mixture Models, group pixels with similar characteristics into distinct segments. They can handle multi-dimensional features but may require careful initialization and parameter tuning.
Deep Learning for Image Segmentation
In recent years, deep learning techniques, particularly convolutional neural networks (CNNs), have revolutionized the field of image segmentation. CNNs can automatically learn hierarchical features from raw image data, making them highly effective in capturing complex patterns and structures.
One popular architecture for medical image segmentation is the U-Net, which consists of an encoder-decoder structure with skip connections. The encoder progressively downsamples the input image to capture contextual information, while the decoder upsamples and combines features to produce a high-resolution segmentation map. Skip connections allow the network to retain spatial information and improve the segmentation accuracy.
Challenges in Medical Image Segmentation
Despite the advancements in image segmentation techniques, there are still several challenges specific to medical images:
- Variability in Anatomy: Human anatomy can vary significantly across individuals, making it difficult to develop generalizable segmentation models.
- Image Artifacts: Medical images may contain artifacts such as noise, motion blur, or intensity inhomogeneity, which can affect the segmentation quality.
- Class Imbalance: In many medical segmentation tasks, the regions of interest (e.g., tumors) occupy a small portion of the image compared to the background, leading to class imbalance and potential bias in the model.
- Limited Annotated Data: Obtaining large amounts of accurately annotated medical data is time-consuming and requires expertise, making it challenging to train robust segmentation models.
In our project, we tackled these challenges by leveraging the nnU-Net framework, which is specifically designed for medical image segmentation. The framework automatically adapts to the dataset characteristics and optimizes the model architecture and hyperparameters accordingly. By utilizing transfer learning and data augmentation techniques, we were able to train an effective model despite the limited annotated data available.
Methodology
- Data Collection: We obtained a dataset of 150 volumetric CT scans with associated segmentation labels for kidneys and tumors from the Kidney-Tumor Segmentation 2023 challenge.
- Data Preprocessing: The CT scans and labels were preprocessed to ensure compatibility with the nnU-Net framework, which we utilized for model development. This involved converting the DICOM files into a suitable format and normalizing the image intensities.
- Model Development: We leveraged the power of the nnU-Net framework to train a 3D U-Net model specifically designed for kidney tumor segmentation. The U-Net architecture is well-suited for medical image segmentation tasks due to its ability to capture both local and global context.
- Model Evaluation: The trained model's performance was evaluated using metrics such as the Dice coefficient and visual assessment to ensure its accuracy and reliability. The Dice coefficient measures the overlap between the predicted segmentation and the ground truth labels.
- Visualization: To make the segmentation results accessible and user-friendly for the IMRA staff, we integrated the MANGO (Multi-image Analysis GUI) software, enabling interactive visualization and analysis. MANGO allowed us to view the segmented regions in 3D and assess the quality of the model's predictions.
Code Snippets
Here's a code snippet showcasing the preprocessing step using the PyDICOM library to load DICOM files:
import pydicom
def load_dicom(file_path):
dicom_data = pydicom.dcmread(file_path)
image_data = dicom_data.pixel_array
return image_data
And here's a code snippet demonstrating the evaluation of the model using the Dice coefficient:
import numpy as np
def dice_coefficient(pred, true, threshold=0.5):
pred = np.asarray(pred > threshold).astype(np.bool)
true = np.asarray(true).astype(np.bool)
intersection = np.logical_and(pred, true)
dice = 2.0 * intersection.sum() / (pred.sum() + true.sum())
return dice
Overcoming Challenges
The path to success was not without its obstacles. Finding suitable training data proved to be a significant challenge due to privacy concerns and the scarcity of labeled medical images. However, with the help of the IMRA staff and the discovery of the KiTS23 dataset, we were able to overcome this hurdle.
Another challenge we faced was the limited computational resources. Training deep learning models on large volumetric CT scans demanded high-performance GPUs, which were not readily available. We had to get creative and leverage cloud platforms like Google Colab to access the necessary computing power.
Results and Impact
The developed 3D U-Net model demonstrated remarkable accuracy in segmenting kidney tumors from CT scans. The integration of MANGO software provided an intuitive interface for medical professionals to explore and validate the segmentation results. This automated approach significantly reduced the time and effort required for manual labeling, enabling efficient generation of 3D printable models for surgical training.
The impact of our project extended beyond the walls of IMRA. By streamlining the creation of synthetic tissue models, we contributed to the advancement of surgical training and the potential improvement of patient outcomes. It was incredibly rewarding to know that our work could make a difference in the lives of both surgeons and patients.
Future Enhancements
- Expand the model to segment additional organs and anatomical structures, further enhancing its versatility and applicability in surgical training.
- Incorporate user-friendly tools for interactive refinement of segmentation results, allowing medical professionals to make fine-grained adjustments as needed.
- Develop a seamless pipeline for direct integration with 3D printing software, streamlining the process from segmentation to physical model creation.
Personal Growth and Reflections
This capstone project at IMRA was a transformative experience for me, both professionally and personally. It challenged me to push the boundaries of my knowledge and skills, while also teaching me the importance of collaboration and perseverance in the face of obstacles.
Working alongside the talented and passionate individuals at IMRA was an absolute privilege. Their dedication to advancing medical technology and improving patient care was truly inspiring. I am grateful for the friendships forged and the invaluable lessons learned throughout this journey.
Project Resources
As I reflect on this incredible experience, I am filled with a sense of pride and accomplishment. The knowledge and skills I gained during this project will undoubtedly shape my future endeavors in the field of data science, machine learning and healthcare. I am excited to see how the work we did at IMRA will continue to evolve and make a positive impact in the world of surgical training and patient care.